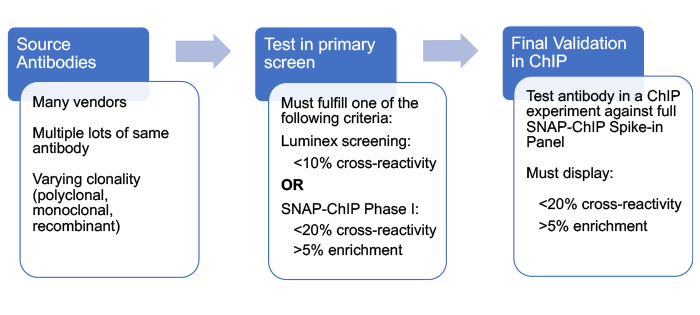
Below is a general outline for the workflow of this project. Briefly, antibodies were profiled in a primary screen designed to rapidly triage antibodies and identify those with high specificity for their target histone post-translational modification (PTM) in a nucleosome context. Selected antibodies were then subjected to rigorous testing using SNAP-ChIP® (Sample Normalization & Antibody Profiling for ChIP) Spike-in Controls, comprising panels of DNA-barcoded recombinant modified designer nucleosomes carrying on- and off-target histone PTMs.
Workflow of This Study:
Here, we provide a brief overview of SNAP-ChIP Spike-ins, our primary antibody screening process, and final antibody validation using SNAP-ChIP Spike-in Panels. The detailed methods can be found at <insert paper link on bioRxiv>. SNAP-ChIP is based on the ICeChIP (Internal Standard Calibrated ChIP) method1, and has been described in a previous paper2.
What are SNAP-ChIP Spike-ins?
SNAP-ChIP Spike-ins are pools of DNA-barcoded recombinant designer nucleosomes carrying distinct histone PTMs. These SNAP-ChIP Panels are spiked into a ChIP reaction, allowing the user to monitor antibody specificity and enrichment on a highly pure, defined nucleosome substrate within the context of a ChIP experiment.
For the data shown on this website we used a native ChIP workflow, in which the SNAP-ChIP Spike-ins were added to chromatin extracts prior to digestion with micrococcal nuclease. Thus, the spike-ins were subjected to the same wash steps and antibody immunoprecipitation (IP) as the experimental sample. Note, the SNAP-ChIP Spike-in Controls are also compatible with crosslinked ChIP; more details can be found here.
The on-target SNAP-ChIP Spike-in was recovered along with sample chromatin bearing the PTM of interest (Figure 1). Following ChIP and DNA isolation, we used qPCR to examine enrichment of each PTM-specific DNA barcode in the SNAP-ChIP Panel. These data were expressed as a percentage relative to the designated on-target PTM, to easily determine the level of cross-reactivity to off-target PTMs. We also performed qPCR on spiked input chromatin to determine enrichment of the PTM target (i.e. efficiency). For the purposes of this website, results are shown for one concentration of antibody (typically 3 µg or 5 µg) tested against SNAP-ChIP Spike-ins added to chromatin from K562 cells (3-10 µg). For more explanation on antibody concentration in ChIP experiments, see our FAQ page.
Importantly, the DNA barcodes can also be detected by next-generation sequencing (NGS), which provides information on antibody specificity, efficiency, and can be used to normalize ChIP-seq data. To learn more about ChIP-seq normalization using SNAP-ChIP Controls, see the SNAP-ChIP Spike-in Controls manual.
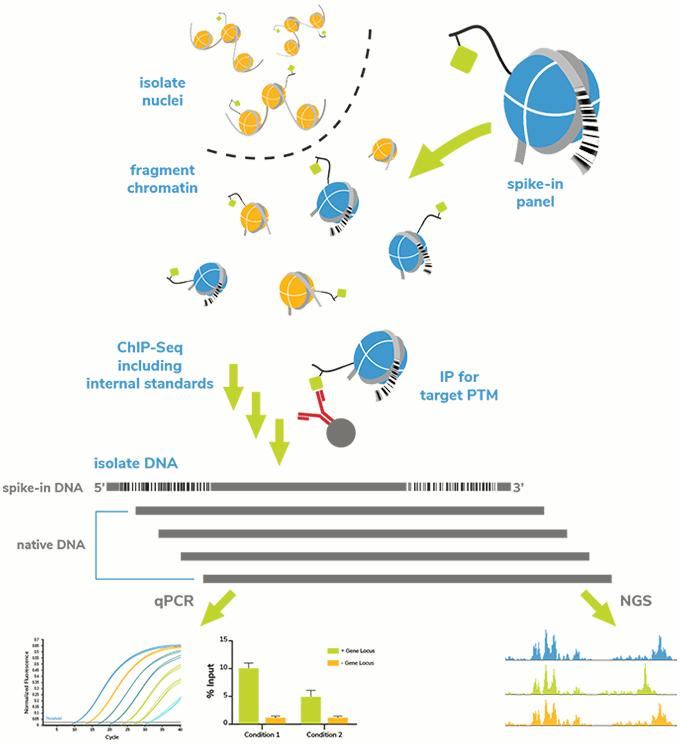
Figure 1: SNAP-ChIP Spike-in workflow. A pool of recombinant nucleosomes with defined PTMs identified by unique DNA barcodes was added to sample chromatin prior to IP. Capture and detection of the barcoded nucleosomes (on- and off-target) provided information about antibody performance (specificity and target enrichment) and technical variability between assays.
Assembly of SNAP-ChIP Spike-ins
SNAP-ChIP Spike-in nucleosomes were developed using recombinant human histones expressed in E. coli. Modifications were added to histone proteins using a proprietary semi-synthetic method, and resulting histones were validated by analytical HPLC and high-resolution mass spectrometry prior to octamer and nucleosome assembly.
Each nucleosome in a SNAP-ChIP panel was wrapped with DNA containing:
- The Widom 601 nucleosome positioning sequence3
- An internal barcode specific to the SNAP-ChIP Panel
- An internal barcode unique to the histone PTM
The barcode sequences were designed to be absent from the human, mouse, fruit fly and budding yeast reference genomes, and thus easily distinguished in qPCR and sequencing. The full sequence of each barcoded DNA is available from the SNAP-ChIP Spike-in product page (www.epicypher.com).
Each nucleosome was quality validated by native PAGE to confirm the absence of free DNA, and equal stoichiometry of histones was determined by SDS-PAGE and Coomassie staining. Histone PTM identity was further confirmed by immunoblot with a highly specific antibody.
The resulting DNA barcoded nucleosomes were combined at equimolar ratios to form distinct SNAP-ChIP Panels, as outlined below.
SNAP-ChIP Spike-in Nucleosome Panels
Antibodies tested in this effort were profiled against a panel of on- and off-target recombinant modified designer nucleosomes, termed SNAP-ChIP Spike-in Panels. These panels were rationally designed to include the most well-studied histone PTMs and highly related marks that are expected to exhibit cross-reactivity.
We currently have three SNAP-ChIP Panels for antibody screening, although we have additional sets in our R&D pipeline.
- KMet-Stat Panel: A set of 15 nucleosomes carrying distinct methyl-lysine PTMs, plus an unmodified nucleosome control.
K-MetStat Panel |
||||
H3K4me1 |
H3K9me1 |
H3K27me1 |
H3K36me1 |
H4K20me1 |
H3K4me2 |
H3K9me2 |
H3K27me2 |
H3K36me2 |
H4K20me2 |
H3K4me3 |
H3K9me3 |
H3K27me3 |
H3K36me3 |
H4K20me3 |
- K-AcylStat Panel: A set of 22 nucleosomes with defined lysine acyl marks, including acetylation, crotonylation, and butyrylation. This panel contains four nucleosomes with combinatorial PTMs, including H3K27 acetylation + H3S28 phosphorylation. H3S28ph is known to alter antibody binding to H3K27ac in some cases4, 5, and provided a useful control for H3K27ac antibody testing. This panel also contains an unmodified recombinant nucleosome as a negative control.
K-AcylStat Panel |
|||
H3 Acetyl PTMs |
H4 Acetyl PTMs |
Non-acetyl PTMs |
Combinatorial PTMs |
H3K4ac |
H4K5ac |
H3K9bu |
H3K27ac + S28ph |
H3K9ac |
H4K8ac |
H3K9cr |
H3K4, K9, K14, K18ac |
H3K14ac |
H4K12ac |
H3K18bu |
H4K5, K8, K12, K16ac |
H3K18ac |
H4K16ac |
H3K18cr |
H2AK5, K8, K13, K15ac |
H3K23ac |
H4K20ac |
H3K27bu |
|
H3K27ac |
H3K27cr |
||
H3K36ac |
|||
- OncoStat Panel: A set of 8 nucleosomes, assembled using H3.3 histones with known oncogenic mutations. H3.3 is included as a control.
OncoStat Panel |
|||
H3.3 |
H3.3K4M |
H3.3K9M |
H3.3K27M |
H3.3G34R |
H3.3G34V |
H3.3G34W |
H3.3K36M |
- Others coming soon: We are developing additional panels targeting other families of PTMs (e.g. arginine methylation) and combinatorial PTMs. This website will be updated as new data are available.
Primary Screening : Triage of Candidate Antibodies
Testing our entire library of histone PTM antibodies using SNAP-ChIP Controls would be a time-intensive and costly project. In addition, given our previous study of H3K4 methyl-state antibodies suggesting >80% of antibodies fail to specifically enrich their target2, we were concerned that such a project would result in mostly negative data. Thus, we developed several nucleosome-based strategies to rapidly screen and identify high-quality antibodies for final testing with SNAP-ChIP Spike-ins.
Luminex® for Rapid, Multiplexed Antibody Screening
We recently developed a new primary screening strategy that combines Luminex xMAP® multiplexing technology with EpiCypher modified recombinant designer nucleosomes for high-throughput specificity testing of histone PTM antibodies. For this method we use MagPlex® Microspheres, sets of carboxylated polystyrene beads that carry unique spectral barcodes which can be detected and quantified on the Luminex FLEXMAP 3D®.
In our approach, sets of barcoded, streptavidin coated Luminex MagPlex beads were coupled to biotinylated recombinant nucleosomes carrying distinct histone PTMs, thus allowing the MagPlex barcode to denote the PTM. The beads were pooled at equal ratios, incubated with a histone PTM antibody (1:250, 1:1000, and 1:4000), and detected on the FLEXMAP 3D using a species-specific anti-IgG antibody coupled to phycoerythrin (PE).
The FLEXMAP 3D provides antibody binding data for a full panel of nucleosomes using a single reaction volume and is compatible with 96- and 384 well plates for improved throughput. Results were expressed relative to the on-target histone PTM. For the purposes of this website, we have chosen to display the results for the lowest antibody concentration that generates the greatest on-target specificity.
In order to progress to final testing in ChIP, with the full panel of SNAP-ChIP Spike-ins, antibodies had to display <10% cross-reactivity to off-target modifications. Notably, we optimized our Luminex screening protocols to be compatible with the stringent wash buffers used in ChIP, making the binding data analogous to that from SNAP-ChIP Spike-in testing.
However, for gold-standard ChIP antibody validation, final screening against SNAP-ChIP Spike-ins in a ChIP experiment was still a crucial step, as Luminex:
- Cannot provide any data on antibody enrichment / efficiency. This information can only be attained using SNAP-ChIP Spike-ins in a ChIP assay.
- Is not reflective of the ChIP experiment; i.e. antibodies are testing in a buffering system, in absence of excess chromatin.
- Does not 100% correlate with results using SNAP-ChIP Spike-ins. In general, antibodies that fail Luminex testing will also fail in ChIP against SNAP-ChIP Controls; however, we found many antibodies that passed Luminex but ultimately failed testing against SNAP-ChIP Spike-ins.
Thus, the best way to determine antibody binding activity in a given application is to test the antibody against defined on- and off-target controls, in the context of the desired experiment. SNAP-ChIP Spike-ins are the only method that meet this rigorous testing standard.
SNAP-ChIP Phase I : Antibody Testing with Subsets of SNAP-ChIP Spike-ins
Our original triage method was a modified version of testing with SNAP-ChIP Spike-ins, in which we profiled antibodies against a small subset of SNAP-ChIP Controls. This Phase I screening focused on closely related PTMs, which tend to show the most cross-reactivity2. For instance, using this approach, H3K4me3 antibodies were only profiled against unmodified, H3K4me1, H3K4me2, and H3K4me3 DNA barcoded nucleosomes, instead of the entire K-MetStat panel. Antibodies that displayed <20% cross-reactivity with off-target PTMs and>5% enrichment in this first screen were then tested against the full SNAP-ChIP panel.
However, this approach still required a significant amount of designer nucleosomes and hands-on time. Thus, once we optimized and validated our Luminex method (above), we transferred preliminary antibody screening to the Luminex platform.
Final Criteria to Pass Phase I Testing |
||
Phase I Assay |
Specificity Metric |
Enrichment Metrics |
Luminex |
<10% cross reactivity to off-target PTMs |
Not tested |
OR |
||
SNAP-ChIP Phase I |
<20% cross reactivity to off-target PTMs |
>5% enrichment vs. input |
Secondary Screen : Certification of Antibodies using SNAP-ChIP Panels
Antibodies that passed triage testing were tested in a final screen, using the appropriate SNAP-ChIP Spike-in Panel spiked into a ChIP experiment (described above). These panels are composed of both on- and off-target PTMs, providing comprehensive analysis of antibody cross-reactivity.
Experiments were performed as previously described1, 2. Specificity and enrichment were determined by qPCR for each spike-in barcode (or by NGS analysis of barcodes). Results are shown for one concentration of antibody (typically 3 µg or 5 µg) tested against SNAP-ChIP Spike-ins added to chromatin from K562 cells (3-10 µg). For more explanation on antibody concentration, see our FAQ page.
Antibodies were considered SNAP-ChIP Certified when they displayed <20% cross-reactivity, relative to on-target PTM, and >5% enrichment vs. Input.
Metrics for Testing Against SNAP-ChIP Spike-ins
Highly specific antibodies exhibit low recovery of off-target PTM barcodes by qPCR (<20%) and high enrichment vs. Input (>5%). These metrics were established as a result of our 2018 Molecular Cell study2, as well as the studies described in our latest paper (ADD REF TO NEW PAPER).
Figure 2 provides an example of a highly specific H3K4me3 antibody vs. an antibody with significant cross-reactivity. Resulting ChIP and ChIP-seq data generated with the nonspecific antibody (Figure 2, left) comprise a mixture of true H3K4me3 signal and contaminating H3K4me2 signal, complicating analysis and leading to incorrect biological interpretations2. Thus, SNAP-ChIP Spike-ins provide an important in-application control for testing antibody specificity against a substrate that replicates the nucleosome context in ChIP. For more on data interpretation, click here.
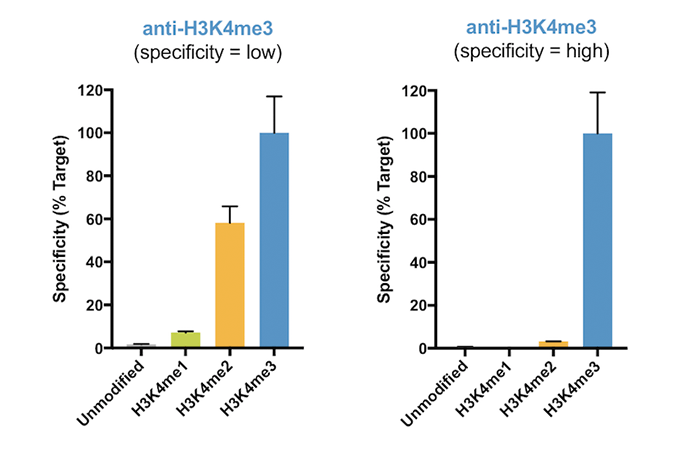
Figure 2: SNAP-ChIP Spike-in profiling data for H3K4me3 antibodies with low specificity (left graph) and high specificity (right graph). The low-specificity H3K4me3 antibody displayed substantial cross-reactivity for H3K4me2. Data are expressed relative to H3K4me3 binding, and sourced from Shah et al.
Antibody sourcing
The antibodies tested in this project came from many vendors, and all antibodies were given a unique EpiCypher ID number to enable blinded, unbiased testing in our assays. However, there are some facts about our antibodies that we want to specify:
- We sourced several types of antibodies for this project, including monoclonal (MC), polyclonal (PC), and recombinant polyclonal (RPC) antibodies. For each antibody we have indicated clonality and clone # (where available), as well as vendor name, catalog number, and lot number.
- We sourced multiple lots of the same antibody (when available) to test lot-to-lot variability in antibody performance. For more on this analysis, see Data Interpretation.
- This project, including the purchase of antibodies and development of testing methods, was funded by the NIH SBIR Grants Program: R44HG008907 (NHGRI); R44GM116584 (NIGMS); R44GM136172 (NIGMS).
- We have recently entered into a partnership with Thermo Fisher Scientific (see our press release) to identify and develop highly specific histone PTM antibodies using SNAP-ChIP Spike-in technology. Future updates to this website will include lot testing of commercially available Thermo Fisher antibodies. Notably, antibodies tested as part of this collaboration undergo the same rigorous testing as those from other companies.
REFERENCES
- Grzybowski AT et al. Calibrating ChIP-Seq with Nucleosomal Internal Standards to Measure Histone Modification Density Genome Wide. Mol Cell 58, 886-99 (2015). PMID:26004229
- Shah RN et al. Examining the Roles of H3K4 Methylation States with Systematically Characterized Antibodies. Mol Cell 72, 162-77 e7 (2018). PMID:30244833
- Lowary PT et al. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol 276, 19-42 (1998). PMID:9514715
- Lau PN et al. Histone code pathway involving H3 S28 phosphorylation and K27 acetylation activates transcription and antagonizes polycomb silencing. Proc Natl Acad Sci U S A 108, 2801-6 (2011). PMID:21282660
- Bock I et al. Detailed specificity analysis of antibodies binding to modified histone tails with peptide arrays. Epigenetics 6, 256-63 (2011). PMID:20962581