Nonspecific Antibodies to Histone PTMs and the Reproducibility Crisis
Antibodies are one of the most widely used tools in all of biomedical research. However, their accuracy has come under intense scrutiny over the past decade, as an alarming number of antibodies fail to specifically recognize their intended target1-3.
This so-called “reproducibility crisis”1 is particularly relevant to the epigenetics community, which relies on antibodies to map the localization of distinct histone post-translational modifications (PTMs) and other factors on chromatin, using chromatin immunoprecipitation (ChIP-seq) or other methods (e.g. CUT&RUN). This information is crucial to understanding the role of chromatin structure in various cellular processes, as well as disease development. Thus, the specificity of antibodies to these targets is incredibly important.
However, antibodies to histone PTMs have been challenging to accurately validate, especially for ChIP-seq4-12. There are several factors that may be contributing to this ongoing problem:
- Use of Histone Peptide Arrays to Validate Antibodies: Modified histone peptide arrays are the established, “gold-standard” method to determine antibody binding specificity to histone PTMs (Figure 1). These assays are inexpensive, fast, and enable profiling against hundreds of single and combinatorial PTMs in a single screen. However, modified histone peptides lack the nucleosomal context present in ChIP-seq experiments and are not subjected to the same stringent wash conditions used in ChIP. As the field continues to learn more about the dynamic interplay between histone tails and DNA, and the impact of various chromatin modifications13-15, it has become clear that a linear peptide may not provide the ideal substrate for testing histone PTM antibody binding.
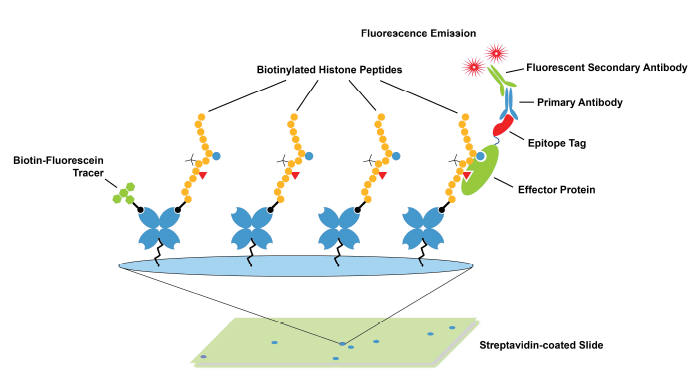
Figure 1: Schematic of histone peptide array. These arrays are commonly used to validate antibodies targeting histone PTMs.
- Antibodies Are Not Always Rigorously Tested: ChIP is a complex and time-consuming experiment, requiring multiple rounds of optimization to perfect chromatin isolation, sonication, buffering conditions, and the immunoprecipitation (IP) process. As a result, antibodies are often not rigorously tested by users prior to using them in a ChIP-seq experiment. Typically, scientists identify an antibody from a highly regarded or cited publication and purchase the most recent lot of this antibody for new experiments.
The result of these customary practices has led to one main outcome: ChIP-seq data are highly variable! The goal of this study was to:
- Develop standardized controls to identify highly specific anti-histone PTM antibodies for ChIP and ChIP-seq experiments.
- Apply these controls to survey the current state of antibodies to histone PTMs.
This website serves as a repository for the resulting data and will be continuously updated as we test additional antibodies.
SNAP-ChIP® Spike-in Controls Demonstrate that Nucleosomes are Key to Histone PTM Antibody Validation in ChIP
Generating accurate profiles of histone PTMs by in ChIP-seq requires highly specific antibodies. Due to the questions surrounding the accuracy of histone peptide arrays, we decided to create a platform that directly examines the specificity of antibodies on a physiological chromatin substrate.
To this end, we developed SNAP-ChIP® (Sample Normalization and Antibody Profiling for ChIP) Spike-in Controls, based on the previously published ICeChIP (Internally Standard Calibrated ChIP) approach16. SNAP-ChIP Spike-ins are composed of panels of DNA-barcoded modified nucleosomes, representing groups of commonly studied histone PTMs, such as methyl-lysine marks (see Methodology). Each DNA barcode denotes a unique histone modification in the spike-in panel.
SNAP-ChIP Spike-in Panels are added to a ChIP experiment prior to IP (see Figure 2). Following IP, recovery of DNA barcodes can be detected by qPCR or next-generation sequencing (NGS) as described previously17, giving a direct readout of specificity (percent on- and off-target DNA barcodes) and enrichment of the target PTM (vs. spiked input chromatin; also termed efficiency).

Figure 2: SNAP-ChIP Spike-in workflow.
Histone Peptide Arrays Do Not Predict Antibody Performance in ChIP
In a previous study17, we performed a head-to-head comparison of SNAP-ChIP Spike-in Controls and standard histone peptide array testing, profiling >50 antibodies to H3K4me0, me1, me2, and me3 using both methods. Remarkably, there was no correlation between antibody performance using SNAP-ChIP Controls and histone peptide arrays (Figure 3). In fact, binding to histone peptides failed to predict performance of antibodies in ChIP-seq experiments!
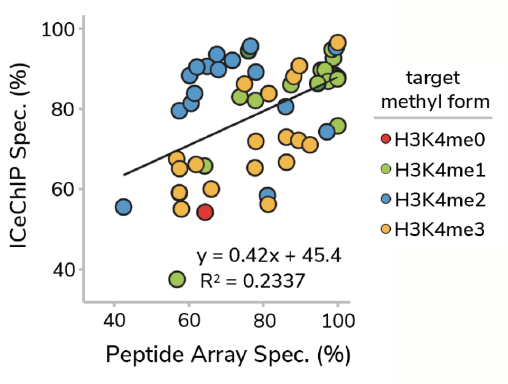
Figure 3: Correlation of antibody binding specificity using histone peptide array (x-axis) vs SNAP-ChIP Spike-ins (y-axis). Percent specificity was calculated as the percent on-target normalized to sum of all H3K4 enrichment. Data sourced from Shah et al.
This surprising result is most easily explained through comparison of histone peptide and nucleosome substrates. As noted above, the linear epitopes represented in modified histone peptides are substantially different from the complex nucleosome structure an antibody actually interacts with in ChIP experiments.
Many “ChIP-grade” Histone PTM Antibodies Display Off-Target Binding
However, another important conclusion from our study is that many commonly used “ChIP-grade” antibodies are not specific for their intended histone PTM target (Figure 4).
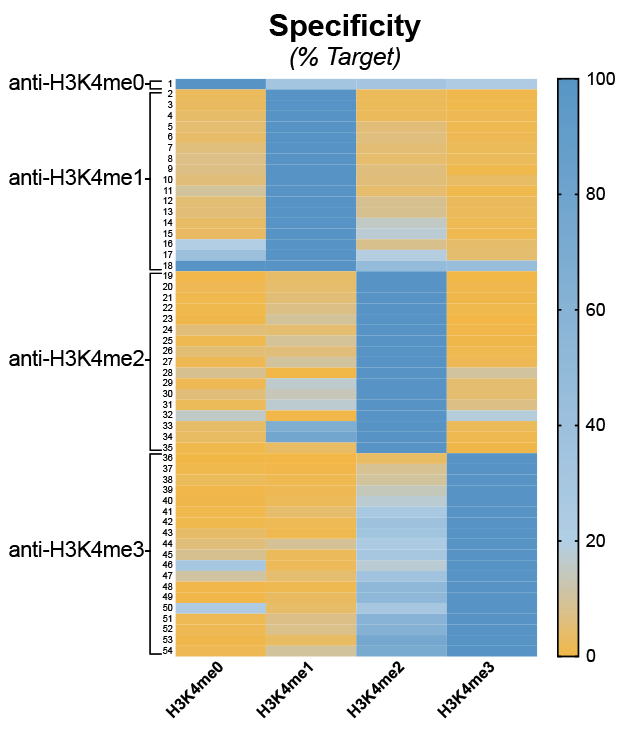
Figure 4: Heatmap summary of H3K4me-state antibody specificity using SNAP-ChIP Spike-ins. A total of 54 antibodies were tested. Binding is shown as a gradient from low (orange) to high (dark blue). Data sourced from Shah et al.
Importantly, we were able to identify a highly specific and efficient (i.e. enrichment from input) antibody for nearly every PTM tested, indicating that:
- Good antibodies to histone PTMs ARE available.
- In-application testing against a defined nucleosome substrate, such as SNAP-ChIP Spike-in Controls, is the ideal strategy to identify highly specific histone PTM antibodies.
The Consequences of Poor Antibody Validation
While analyzing our H3K4me antibody specificity data (Figure 4, above), we were particularly dismayed by the results for H3K4me3 antibodies. In fact, of the 19 H3K4me3 antibodies tested using SNAP-ChIP Controls, 16 exhibited >20% cross-reactivity with H3K4me2 (see Figure 4)17.
This was especially alarming, as H3K4me2 is 3-5 times more abundant in the cell compared to H3K4me318. In addition, this low-specificity antibody group included many of the most highly cited and widely used H3K4me3 antibodies. Thus, much of the binding attributed to H3K4me3 in previous studies may actually be contaminating H3K4me2 signal!
To test this theory, we performed ChIP-seq experiments using three different antibodies17 (Figure 5):
- A H3K4me2 antibody with high specificity.
- A H3K4me3 antibody with binding to both me3 and me2 marks, that has >1,000 citations.
- A H3K4me3 antibody that only binds me3.
Comparison of ChIP tracks (Figure 5) revealed that use of a low specificity H3K4me3 antibody resulted in significant contaminating signal from H3K4me2.
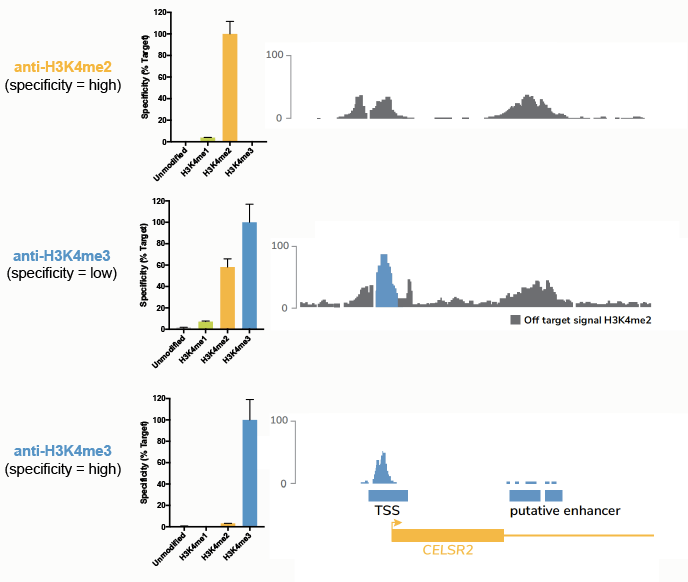
Figure 5: Comparison of ChIP-seq tracks using a panel of antibodies tested against SNAP-ChIP Spike-ins. The top panel shows specificity data from SNAP-ChIP Controls (left) and ChIP-seq tracks (right) for a highly specific antibody targeting H3K4me2. The middle and bottom panels show the same data for low-specificity and high-specificity anti-H3K4me3 antibodies, respectively. Data sourced from Shah et al.
This finding has important biological implications. In addition to the classical association of H3K4me3 with active promoters19-24, H3K4me3 has also been described at actively transcribed enhancers25, 26, and “broad domains” of low-level H3K4me3 enrichments have been associated with expression of genes dictating cell identity27.
However, after correcting for background H3K4me2 and me1 signal using SNAP-ChIP Spike-ins, none of these results were replicated17. It is likely that some of the biological functions assigned to H3K4me3 are due to the use of poorly validated antibodies.
Thus, it is essential to properly validate histone PTM antibodies against a nucleosome substrate in the context of a ChIP experiment. Of note, SNAP-ChIP Spike-in Controls are the first and only technology that allow you to examine antibody specificity during your ChIP experiment.
Mounting Evidence Suggests Widespread Problems with Histone PTM Antibodies
We were curious: is this poor specificity in ChIP a common problem among all histone PTM antibodies, or isolated to a few PTMs or manufacturers? Following publication of our H3K4-methylation study in 201817, we developed additional SNAP-ChIP Spike-in Panels targeting other methyl-lysine marks (K-MetStat Panel), as well as a comprehensive panel to profile antibodies against histone lysine acylation (K-AcylStat Panel) and oncogenic histone mutations (OncoStat Panel).
We sourced hundreds of antibodies to this diverse PTM set and began testing.
Our results demonstrate a pervasive problem with histone PTM antibody specificity. This problem is not unique to a certain set of PTMs or companies, suggesting the problem is intrinsic to the standard processes used for developing and validating antibodies for ChIP.
An early heatmap from this project, showing results from profiling antibodies to histone lysine methylation on H3K4, H3K9, H3K27, H3K36 and H4K20, illustrates this problem (Figure 6). For most PTMs, >80% of antibodies tested displayed unacceptable levels of cross-reactivity (>20% binding to off-target PTM) or enrichment (<5%).
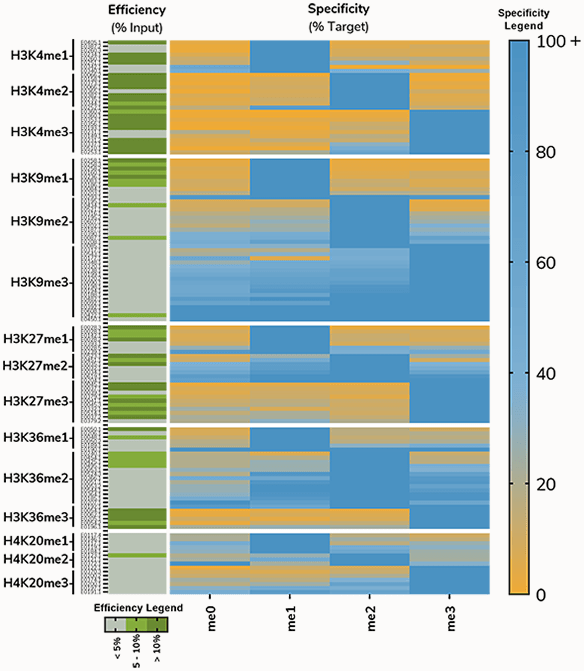
Figure 6: Antibodies to histone methyl-lysine PTMs tested using SNAP-ChIP Spike-ins. Heatmap on right shows specificity relative to on-target PTM; Blue indicates high binding, while orange denotes low binding. Heatmap on the left shows efficiency, or enrichment of on-target nucleosome compared to input. Efficiency is divided into three categories: <5% (gray), 5-10% (light green), >10% (dark green).
Comprehensive Testing to Identify Highly Specific Antibodies
Similar to our 2018 study on H3K4me-state antibodies17, we have been able to identify highly specific and efficient antibodies to most histone PTMs. However, the prevalence of poorly validated “ChIP-grade” antibodies, combined with the demand in the chromatin field for improved tools, led us to start a larger project to identify the best antibodies for ChIP-seq against histone PTMs.
This website is the result of that effort and will continue to grow for the foreseeable future as new antibodies become available for testing, and as we develop additional SNAP-ChIP Spike-in Panels.
We are actively expanding the histone PTM targets profiled by our study; if your histone PTM or antibody of choice isn’t listed on this website, be sure to contact us. Our R&D team is constantly sourcing new antibodies and developing novel SNAP-ChIP Controls, all to ensure scientists have best-in-class tools for their chromatin profiling experiments.
REFERENCES
- Baker M. Reproducibility crisis: Blame it on the antibodies. Nature 521, 274-6 (2015). PMID:25993940
- Bordeaux J et al. Antibody validation. Biotechniques 48, 197-209 (2010). PMID:20359301
- Bradbury A et al. Reproducibility: Standardize antibodies used in research. Nature 518, 27-9 (2015). PMID:25652980
- Bock I et al. Detailed specificity analysis of antibodies binding to modified histone tails with peptide arrays. Epigenetics 6, 256-63 (2011). PMID:20962581
- Egelhofer TA et al. An assessment of histone-modification antibody quality. Nat Struct Mol Biol 18, 91-3 (2011). PMID:21131980
- Fuchs SM et al. Influence of combinatorial histone modifications on antibody and effector protein recognition. Curr Biol 21, 53-8 (2011). PMID:21167713
- Fuchs SM et al. Antibody recognition of histone post-translational modifications: emerging issues and future prospects. Epigenomics 3, 247-9 (2011). PMID:22122332
- Goens G et al. Characterization and quality control of antibodies used in ChIP assays. Methods Mol Biol 567, 27-43 (2009). PMID:19588083
- Nishikori S et al. Broad ranges of affinity and specificity of anti-histone antibodies revealed by a quantitative peptide immunoprecipitation assay. J Mol Biol 424, 391-9 (2012). PMID:23041298
- Peach SE et al. Quantitative assessment of chromatin immunoprecipitation grade antibodies directed against histone modifications reveals patterns of co-occurring marks on histone protein molecules. Mol Cell Proteomics 11, 128-37 (2012). PMID:22442256
- Rothbart SB et al. An Interactive Database for the Assessment of Histone Antibody Specificity. Mol Cell 59, 502-11 (2015). PMID:26212453
- Rothbart SB et al. Poly-acetylated chromatin signatures are preferred epitopes for site-specific histone H4 acetyl antibodies. Sci Rep 2, 489 (2012). PMID:22761995
- Morrison EA et al. The conformation of the histone H3 tail inhibits association of the BPTF PHD finger with the nucleosome. Elife 7, (2018). PMID:29648537
- Morrison EA et al. DNA binding drives the association of BRG1/hBRM bromodomains with nucleosomes. Nat Commun 8, 16080 (2017). PMID:28706277
- Weaver TM et al. Reading More than Histones: The Prevalence of Nucleic Acid Binding among Reader Domains. Molecules 23, (2018). PMID:30322003
- Grzybowski AT et al. Calibrating ChIP-Seq with Nucleosomal Internal Standards to Measure Histone Modification Density Genome Wide. Mol Cell 58, 886-99 (2015). PMID:26004229
- Shah RN et al. Examining the Roles of H3K4 Methylation States with Systematically Characterized Antibodies. Mol Cell 72, 162-77 e7 (2018). PMID:30244833
- Leroy G et al. A quantitative atlas of histone modification signatures from human cancer cells. Epigenetics Chromatin 6, 20 (2013). PMID:23826629
- Guenther MG et al. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77-88 (2007). PMID:17632057
- Santos-Rosa H et al. Active genes are tri-methylated at K4 of histone H3. Nature 419, 407-11 (2002). PMID:12353038
- Schubeler D et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev 18, 1263-71 (2004). PMID:15175259
- Wysocka J et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442, 86-90 (2006). PMID:16728976
- Vermeulen M et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131, 58-69 (2007). PMID:17884155
- Lauberth SM et al. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell 152, 1021-36 (2013). PMID:23452851
- Pekowska A et al. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J 30, 4198-210 (2011). PMID:21847099
- Core LJ et al. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet 46, 1311-20 (2014). PMID:25383968
- Benayoun BA et al. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell 158, 673-88 (2014). PMID:25083876