1. How much do antibodies change lot-to-lot?
Antibodies can display drastic changes in specificity and efficiency between production lots. Figure 1 uses different lots of the same polyclonal H3K27me1 antibody as a representative example. To make lot-specificity data easier to navigate, on each individual antibody page we have included a table that lists all lots that have been tested using Luminex and/or SNAP-ChIP Spike-ins.
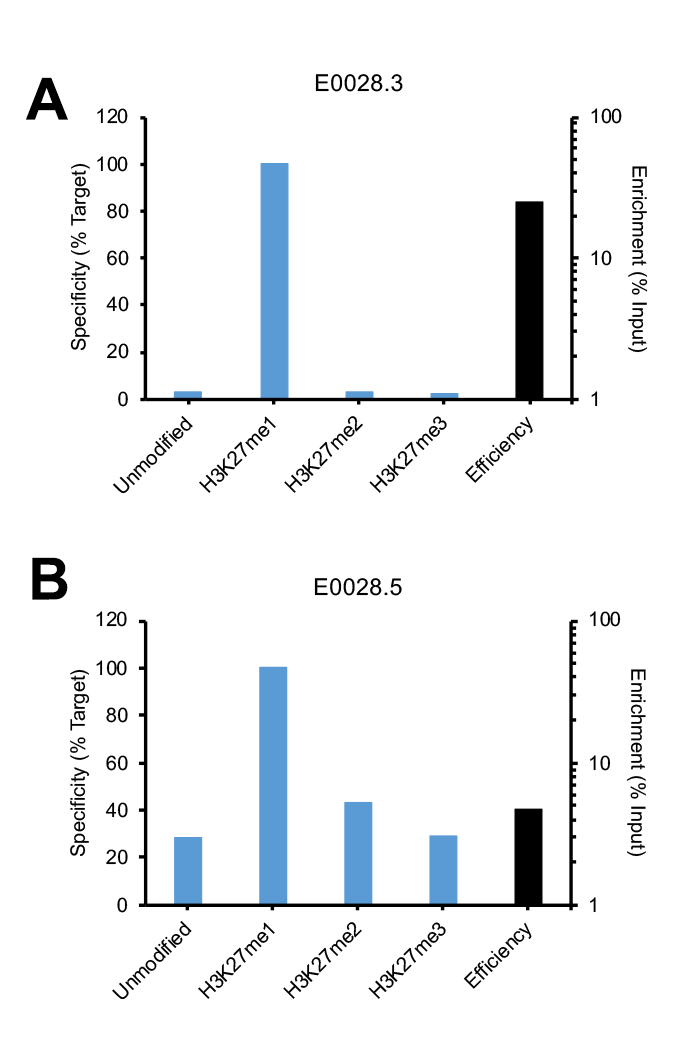
Figure 1: SNAP-ChIP Spike-in testing of different lots of the same polyclonal H3K27me1 antibody (E0028). (A) Lot 3 met SNAP-ChIP testing metrics, with <20% cross reactivity >5% enrichment of H3K27me1 vs. input chromatin, while (B) Lot 5 failed to meet these same metrics.
This problem is widespread and unpredictable, and occurs irrespective of histone PTM target, antibody production method, and vendor (see Interpretation). Monoclonal antibodies are traditionally thought to be more consistent across lots compared to polyclonal antibodies, which are widely understood to exhibit variability. However, even monoclonal antibodies can exhibit drastic differences across lots (see Figure 2 for a representative example). Most commonly, lot-to-lot differences manifest as a result of changes in antibody concentration, which primarily affect antibody efficiency and signal-to-noise. However, monoclonal antibody specificity can also change from lot-to-lot. Thus, it is critical to test every new lot of antibody prior to using it in an experiment.
2. If I dilute my antibody, will it improve specificity?
Probably not. We have conducted extensive experiments to assess how antibody specificity is affected by serial dilutions in both Luminex and SNAP-ChIP Spike-in experiments. Rarely does diluting the antibody dramatically improve specificity; however, there are rare cases where this has been observed (see Figure 3). In the majority of cases, antibody efficiency in ChIP is decreased with lower amounts of antibody, but specificity is largely unaffected (Figure 3).
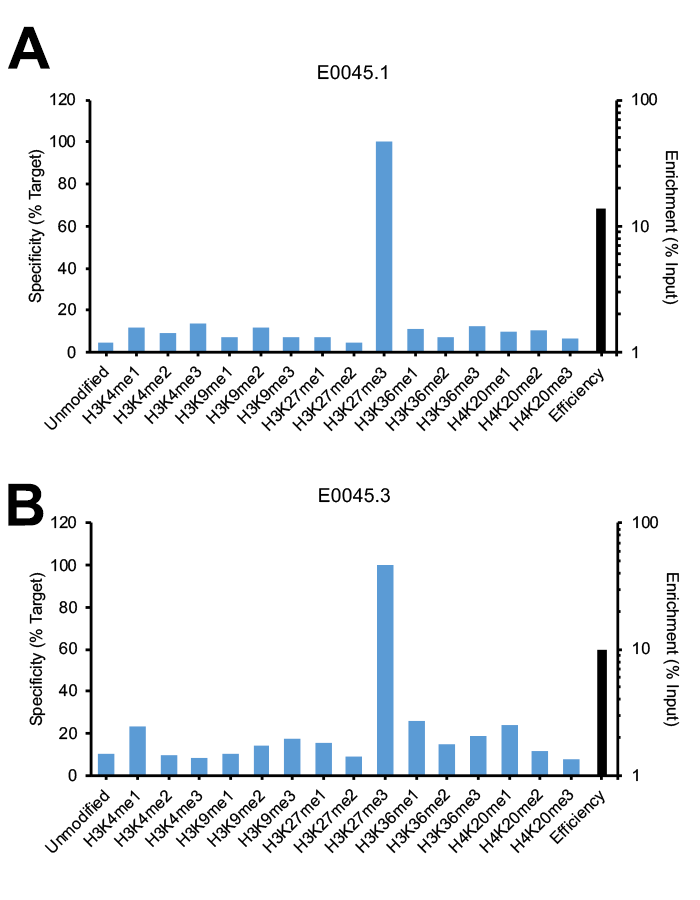
Figure 2: SNAP-ChIP Spike-in testing of different lots of the same monoclonal H3K27me3 antibody (E0045). (A) Lot 1 met SNAP-ChIP testing metrics, with <20% cross reactivity >5% enrichment of H3K27me3 vs. input chromatin, while (B) Lot 3 failed to meet these same metrics.
<Figure 3 will be inserted here, I don’t have them at this time…>
In general, we suggest avoiding antibodies that require dilution to improve specificity, as this may result in greater intra-experimental variability, especially when compared to an antibody that does not exhibit cross-reactivity at any dilution. Additionally, fit-for-purpose reagents have been identified for the vast majority of PTMs studied, which do not require extensive dilutions in order to improve performance.
3. What about specificity in other applications (i.e. western blot, immunostaining)?
SNAP-ChIP Spike-ins were developed to determine antibody specificity in a ChIP experiment, but they may not accurately predict performance in other assays. Although many SNAP-ChIP certified antibodies may produce signal in western blot, immunostaining, or ELISA applications, we do not suggest using SNAP-ChIP Panels as a validation tool for these experiments. Indeed, our work with SNAP-ChIP Spike-in technology has demonstrated that histone PTM antibody validation should be performed in the desired application, using a defined substrate (linearized peptide vs. three-dimensional nucleosome) similar to that encountered in the assay.
4. Are polyclonal or monoclonal antibodies better?
It is commonly assumed that monoclonal antibodies are more reliable tools for ChIP-seq and other applications. However, we have observed problems with specificity and efficiency in both monoclonal and polyclonal antibodies, and both formulations can exhibit extreme lot variation (see Question #1). Thus, no matter what type of antibody you choose for your assay, you should validate it using defined spike-in controls, such as SNAP-ChIP Panels.
5. How do I cite this data in my manuscript?
XXXXXX (citation info + link to paper)
6. How does low antibody efficiency impact my experiment?
Antibody efficiency refers to the enrichment of the target SNAP-ChIP spike-in barcode compared to spiked Input chromatin. Thus, it is a direct measurement of how well your antibody enriches for the target histone PTM.
Using a low-efficiency antibody can lead to high levels of background in ChIP-seq, particularly when profiling a histone PTM with low abundance. When the physiological levels of your target PTM are low, using a highly efficient and specific antibody enables improved recovery and signal : noise (see Figure 4).
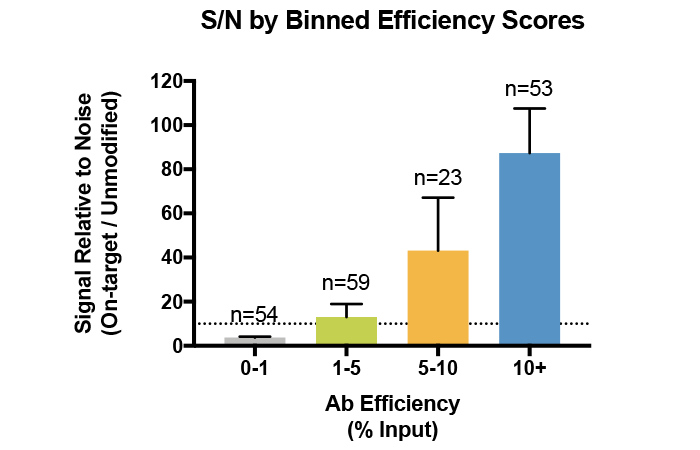
Figure 4: Average signal-to-noise (S/N) for H3K4me0, me1, me2, and me3 antibodies tested against SNAP-ChIP Spike-in Controls. Antibodies are binned by efficiency score, revealing that improved enrichment correlates with improved recovery and S/N. Data sourced from Shah et al. 2018.
7. The SNAP-ChIP Spike-in protocol uses native ChIP. How do these antibodies perform in cross-linked ChIP?
We (and others1, 2) have tested a subset of SNAP-ChIP Certified antibodies in cross-linked ChIP-seq with SNAP-ChIP spike-ins (data not shown). The vast majority perform similarly between the different ChIP protocols. Notably, we modify our approach for cross-linked ChIP experiments, and add SNAP-ChIP Spike-ins after sample fixation and shearing. This is necessary to preserve the spike-ins, which can be damaged by shearing methods, such as sonication. However, adding SNAP-ChIP spike-ins after chromatin fragmentation allows you to control for all subsequent steps of the ChIP-seq experiment.
8. Can I see the full protocol used for the SNAP-ChIP Spike-in experiments?
For our full detailed protocol, we refer you to our 2018 Molecular Cell study and our recent bioRxiv preprint. We have also summarized our approaches in the Methodology section of this website.
9. An antibody that fails the Luminex and/or SNAP-ChIP Spike-in validation gave me good-looking ChIP tracks. What’s the deal?
Our Luminex and SNAP-ChIP Spike-in screening studies are designed to identify highly specific and efficient antibodies with reliable on-target performance in ChIP-seq. It is not meant to select for antibodies that generate a certain peak amplitude or shape. Thus, it is feasible for an antibody that fails SNAP-ChIP or Luminex to generate ChIP-seq data. If this happens with your antibody, it could be for one or more reasons:
- Your data is contaminated by off-target signal. Again, it is entirely possible that your ChIP-seq data are contaminated with off-target peaks. Such data would look completely “normal”, but still be inaccurate; this is why it is crucial to include spike-in controls in every ChIP-seq experiment.
Our previous work3 (and others2) has shown this to be the case for many H3K4me3 antibodies. As shown in Figure 5 (middle panel), a low-specificity H3K4me3 antibody produces apparently normal ChIP-seq peaks. But when compared to ChIP-seq data generated using highly specific, SNAP-ChIP Certified H3K4me2 and H3K4me3 antibodies (top and bottom panels, respectively), it is clear that a substantial amount of the signal from this low-specificity antibody is actually from H3K4me2. See our 2018 Molecular Cell study and Background for more information on this specific example. Importantly, we have observed extensive cross reactivity for antibodies targeting nearly every PTM (insert ref to new paper!), indicating a widespread and unrecognized problem in the field.
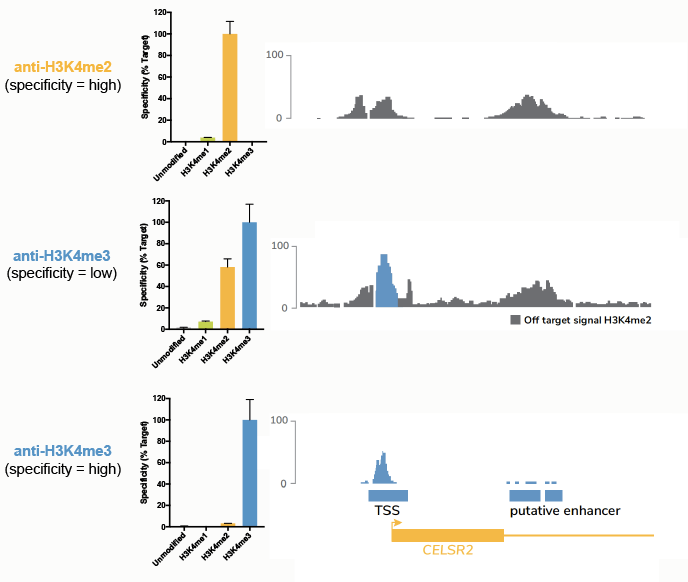
Figure 5: Comparison of ChIP-seq tracks using a panel of antibodies tested against SNAP-ChIP Spike-ins. The top panel shows specificity data from SNAP-ChIP Controls (left) and ChIP-seq tracks (right) for a highly specific antibody targeting H3K4me2. The middle and bottom panels show the same data for low-specificity and high-specificity anti-H3K4me3 antibodies, respectively. Data sourced from Shah et al.
- You are using a different lot number than what was tested in our study. Is this antibody the same lot that was tested in our database? You may be using a different lot of the same antibody, as many antibodies are known to display batch variation in specificity and/or enrichment.
- Under your particular experimental conditions, the antibody performs well. We use native ChIP for our assays, but it is possible that under different conditions an antibody may perform better or worse. NOTE: this is precisely why it is critical to include spike-in controls for every experiment. This way technical variability can be monitored, and any observed cross-reactivity can be used to accurately interpret biological findings.
10. The ChIP antibody that I use is validated by histone peptide arrays. Isn’t that sufficient?
We have shown that histone peptide arrays fail to predict histone PTM antibody binding activity in ChIP (see Background and our 2018 Molecular Cell paper). Histone peptide arrays use modified histone peptides, which do not model the endogenous chromatin structure. In addition, these arrays are not compatible with ChIP wash buffers, and thus fail to replicate experimental conditions.
In order to validate your antibody for ChIP, you need to test the antibody against a defined chromatin substrate in the context of a ChIP-seq experiment. SNAP-ChIP Spike-in panels are the only tool that meet this rigorous standard.
REFERENCES
- Tay RE et al. Hdac3 is an epigenetic inhibitor of the cytotoxicity program in CD8 T cells. J Exp Med 217, (2020). PMID:32374402
- Lam KG et al. Cell-type-specific genomics reveals histone modification dynamics in mammalian meiosis. Nat Commun 10, 3821 (2019). PMID:31444359
- Shah RN et al. Examining the Roles of H3K4 Methylation States with Systematically Characterized Antibodies. Mol Cell 72, 162-77 e7 (2018). PMID:30244833